Six Easier Pieces*:
Discovering the 3D structure of Substance P,
using only free internet tools.
L.
Van Warren
University of Arkansas for Medical Sciences
Department of Anatomy - College of Medicine
& Arkansas Center for Cancer Research
June 12, 1999
Introduction:
Living systems favor the use of small
molecules as tokens of exchange in a complex self-sustaining biochemical economy.
Examples are oxygen, and carbon dioxide in respiration,
the alkali metals in serum electrolytes, glucose
in energy storage and a host of others. There are several neurotransmitters
that are essential for nerve signal transmission in biological organisms. Most
of these are also relatively small molecules. Among them are histamine,
dopamine,
and serotonin.
Others, such as bradykinin
and Substance P are molecular bills of slightly larger denomination
, the later consisting of 11 amino acids linked by 10 peptide bonds. The structure
of the common neurotransmitters have been published on the world wide web, the
exception being the subject of this short note, the tachykinin
called Substance P.
The official definition of tachykinins
from the NCBI database is:
TACHYKININS ARE ACTIVE
PEPTIDES WHICH EXCITE NEURONS, EVOKE BEHAVIORAL RESPONSES, ARE POTENT VASODILATORS
AND SECREATAGOGUES, AND CONTRACT (DIRECTLY OR INDIRECTLY) MANY SMOOTH MUSCLES.
Further investigation
reveals that the term Substance P is an alias for several analogs, whose
amino acid sequence varies with species.
Some of these are given in the following table:
Table 1: Substance
P analogs by species
|
Species
|
Sequence
|
|
|
|
|
|
|
|
|
same
as above
|
|
|
same
as above
|
|
|
|
|
|
same
as above
|
|
|
|
|
|
|
|
|
|
At this writing a thorough search
of the web does not yield the three dimensional configuration of Substance P.
The "P" in substance P actually stands for powder. When isolated for
the first time from horse intestine in the 1930s, it was in powder form.
The point of this technical note
is twofold, first the completion of an errand to determine the 3D structure
of substance P for a cancer research project, and secondly, to show how it is
relatively easy to apply this procedure to other molecules of interest.
Six Easy Steps: Deducing 3D biochemical
structures via the web:
Step 1:
the amino acid sequence is obtained via
NCBI Entrez, it is: KPRPGQFFGL
M as seen in the table above.
Step 2: using an/
or the simpler
quick aa table one can find that this 11 symbol string translates to:
lysine-proline-arginine-proline-glycine-glutamine-phenylalanine-phenylalanine-glycine-leucine-methionine
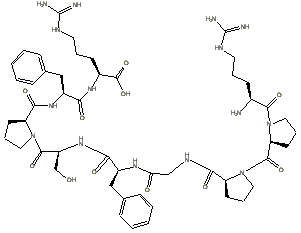
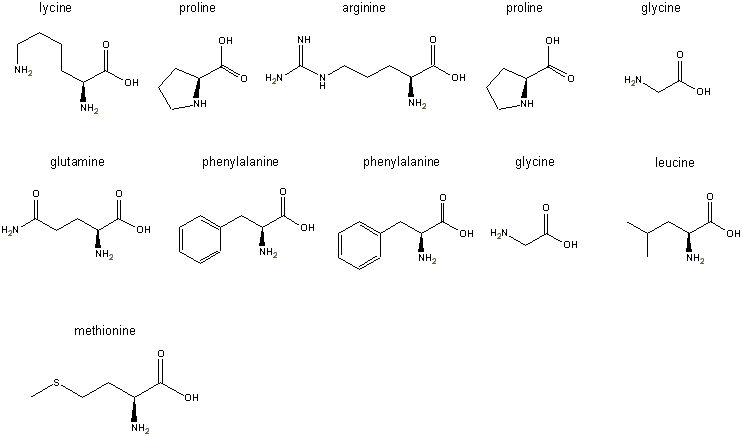
Step 3: the L-forms of all
the amino acids are pasted into the freely distributed ChemDraw
Net product left to right, top to bottom as shown in the figure below:
Step 4: the peptide bonds are formed by condensing out a water using
a hydrogen from the primary amine groups and a hydroxyl group from the carboxyl
groups:
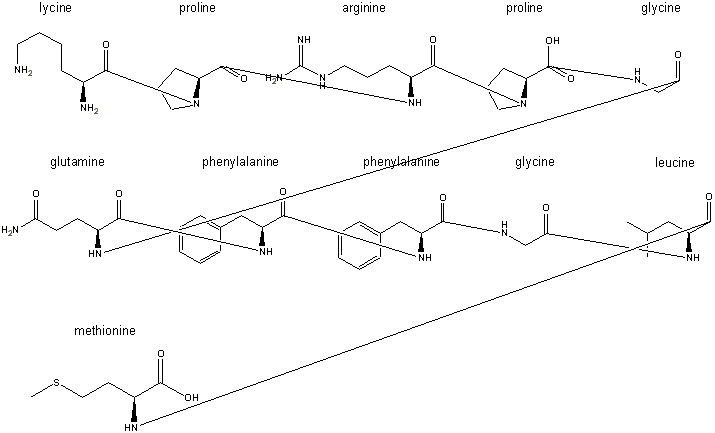
These peptide bonds are formed by
when the line is connected. A hydrogen disappears automatically, but the hydoxyl
groups, such as the one seen remaining on the proline in the first row, are
erased manually after assembly.
Step 5: In the freeChemDraw Net 4.5
tool there is an option called clean-up structure. Clicking on it produces a
more aesthetic 2-D drawing of the structure:
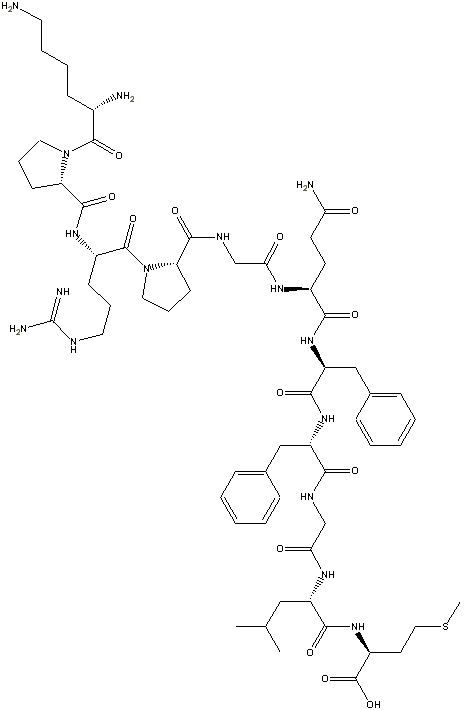
This feature is not in the ChemDraw Standard ($299
academic version), but it is in the ChemDraw Pro version (also $299
at this writing Cambridge software runs specials from time to time, but
they can do their own advertising!). If you install ChemDraw Standard, it
will ask you if you to delete your ChemDraw Net 4.5, which causes structure
clean-up to stop working.
Step 6: In the final
step, we select the entire structure in ChemDraw using either the square or
round marquee lasso tools. We then choose the option Copy As from the
Edit menu and which results in a text representation of the structure
being transferred to the system clipboard in SMILES format. Pasting the clipboard
contents into another application results in this SMILES text string, representing
the structure of Substance P, being transferred. This SMILES text string looks
like:
N[C@H](C([C@H]1[C@@H](CCC1)C(NC(NCCC[C@H](N)C([C@H]2[C@@H]
(CCC2)C(NCC(NC(CC[C@H](N)C(N[C@H](C(N[C@H](C(NCC(N[C@@H]
(CC(C)C)C(N[C@@H](CCSC)C(O)=O)=O)=O)=O)CC4=CC=CC=C4)=O)
CC3=CC=CC=C3)=O)=O)=O)=O)=O)=N)=O)=O)CCCCN
When this text string
is emailed to the Corina
Web Service in Germany, the result is the three dimensional structure below:
The SMILES string for
substance P turns out to be 12 characters too long for the interactive 3D service,
otherwise we could have obtained the structure in almost real time. The overnight
service via email instructions is utilized instead.
Althought the resulting
conformation is useful for appreciating three dimensional molecular structure,
it is non-unique and often unrealisitic. It is necessary to perform energy minimization
to predict plausible configurations of the molecule. This was done using the
CS Chem3D Pro software, which was also available free of charge through a special
CS web promotion:
The conformation of
the final molecule is much different from that of the original. The reader
is urged to enumerate the number of plausible conformations as a combinatorial
exercise.
Conclusion
This simple six step
process can be applied to a large variety of biochemistry applications. The
author intends to continue refining this approach for the illustration of
biochemical
pathways. The enthusiastic reader is urged to repeat these steps for the
other analogs listed in table 1.
Acknowledgments
*Apologies to the late great Richard
Feynman for the title. His book by the same name is wonderful and the title is
a tribute. The tools ChemDraw 3D and
the Corina 3D structure
service have made this otherwise tedious and difficult task nearly effortless.
A debt of gratitude is owed by the biochemistry community to them for the fine
capabilities they are providing. Thanks to Don DeLuca of UAMS and Steven Fleisler
of St. Louis University for clarifying the condensation conventions for peptide
bonds. Thanks to MDL for developing the Chime
plug-in for 3D viewing in Netscape and Explorer. Thanks to Eric
Martz for clarifying Chime installation procedures on the server side. Appreciation
to Dr. Reed Thompson of UAMS/ACRC for sponsoring related
work on cancer pain, which gave rise to this neurotransmitter investigation.
A sincere note of gratitude is due to Dr. Wolf-D. Ihlenfeldt Computer Chemistry
Center, University of Erlangen-Nuernberg Erlangen (Germany) who has developed
an interactive structure service. Appreciation to Marilyn Fulper-Smith for
review and assistance. A special thanks also to Mark
Turpin whose web sponsorship and innovation, none of this would have been
possible. Emily Lowry, a high school intern at UCSF, provided the lexical history
of substance P.